St Anne’s Academic Review 8 – 2018
Emerging Viral Pathogens: New Threats in Global Health
Ciaran Gilbride, Doctoral Training Centre in Interdisciplinary Bioscience
STAAR 8 – October 2018, pp. 37-47
——————————–
Published: 03 Oct 2018
Review process: Open Peer Review
Draft First Uploaded: 22 Aug 2018. See draft and reviewers’ comments.
Abstract
Emerging Viral Pathogens are some of the most deadly disease that humanity have to face, with high mortality rates and the potential to cause mass panic. The Ebola crisis in 2013-2015 and the Zika outbreak of 2016 brought the dangers of emerging pathogens to the public consciousness, as well as earlier crises such as Swine flu, Avian flu and SARS this millennium. Often occurring at the frontiers of humanity, emerging pathogen outbreaks are predominantly zoonotic, jumping from animal hosts into humans with no pre-existing immunity. In this review I introduce the biology of emerging viral diseases, how they infect human hosts, look at case studies on emerging pathogens with pandemic potential and consider HIV, an emerging pathogen which has reached pandemic levels.
Introduction
Emerging pathogens present one of the biggest biological threats to human health. 100 years ago Spanish Flu ravaged the world following WWI, causing illness in up to 30% of the population [1] and killing 100 million; many more fatalities than the preceding four year war. Even further back, the synergy of black death in the old world[2], and smallpox in the new, is considered a contributory factor in the Little Ice Age[3]; lower populations unable to farm sufficient land to prevent forest regrowth and thus CO2 absorption. In the late 20th and early 21st centuries we have seen the outbreak of HIV[4], Ebola[5], SARS[6] and multiple strains of pandemic flu[7-8]; all representing major threats to global health.
The most severe emerging pathogens, and the ones discussed in this review, are often viruses. Viruses are one of the smallest biological entities, consisting of nucleic acid (either RNA or DNA), wrapped in a protein shell called a capsid[9]. Virus structures are so small and their parasitic lifestyle so different to, and dependent on, other organisms that their definition as living remains a scientifically contentious issue[10].
The Viral Life Cycle
The viral life cycle (Fig. 1) begins when a virus adheres to a host cell. The interaction is mediated by two proteins; one on the virus, one on the target cell. The first is the viral glycoprotein, a protein which has been modified by covalently attached sugars; this glycosylation determined by the antecedent host cell [11]. The glycoprotein is unique to each virus species and, exposed on the capsid or viral envelope surface, can interact with the host. To initiate cell entry requires the binding of the viral glycoprotein with a host membrane protein. Each viral glycoprotein interacts with a specific receptor, or set of receptors, displayed on the target cell membrane[11]. The variation in receptor expression across host cell types and between host species determines the tropism (“cell-specificity”) of viral infection[11].
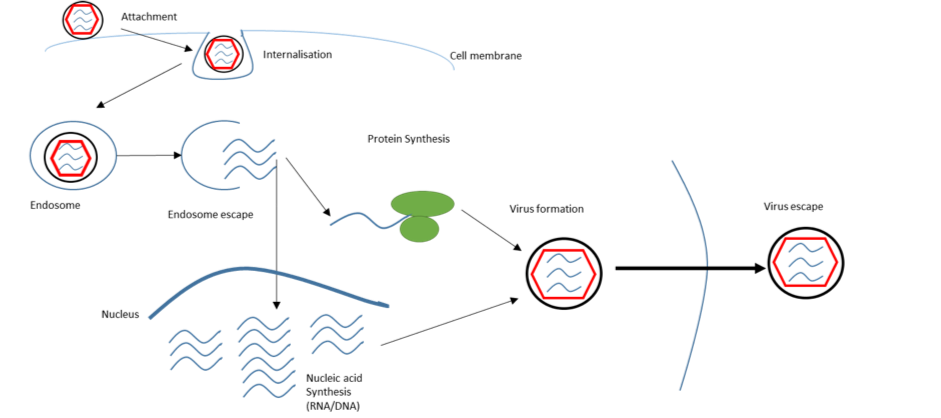
Figure 1 Schematic showing the viral lifecycle of an enveloped virus. Viruses enter cells by a variety of mechanisms. Enveloped viruses attach to the cell surface and are internalised into an endosome; an intracellular membranous compartment. The viral envelope can fuse with the endosomal membrane, releasing the viral genome into the host cell. The host cell machinery then replicates the viral genome and translates its genes into viral proteins. The viral genome and proteins assemble into a capsid, containing the genetic material to produce more viruses in future host cells.
Measles (MeV) is well characterised virus with an elucidated entry mechanism. The surface hemagglutinin is the glycoprotein[12] and can interact with multiple cell surface receptors, such as SLAM and Nectin-4[12]. However, within the same species we can see viral glycoproteins adapt to target new receptors. The hemagglutinin of a MeV laboratory strain has adapted to bind to, and initiate cell entry through, the CD46 protein[12] for example (Fig. 2). The adaption gives the virus a broader target cell tropism, and has been seen in mutant populations outside the lab[12].
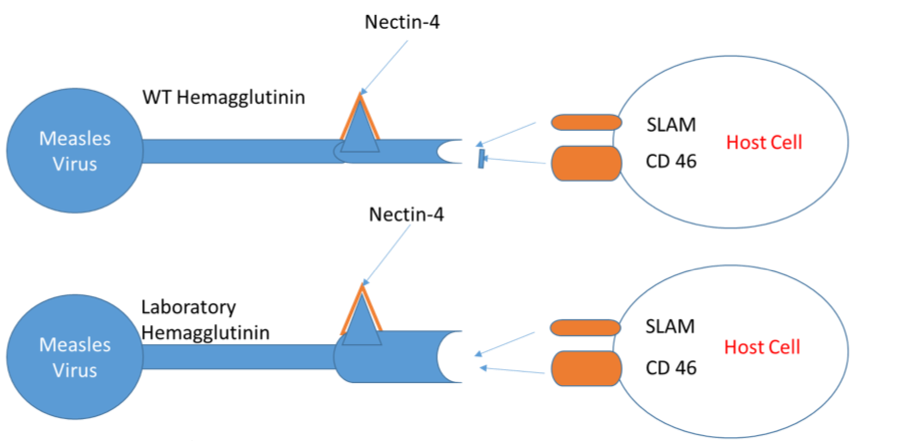
Figure 2 Binding of WT and Laboratory Strain MeV hemagglutinin on the viral envelope to receptors on the host cell surface. The strains differ by a hemagglutinin mutation, allowing the laboratory strain to bind to a wider range of receptors on the host cell surface; hence allowing a wider repertoire of cell types to be targeted within the host.
PPR (Peste des petits ruminants) is a potential emerging pathogen closely related to measles. In small ruminants its symptoms include fever, gastroenteritis and necrotizing stomatitis, culminating in death[13]. The differences lie exclusively in host receptor preference. PPR hemagglutinin, like MeV hemagglutinin, binds to the SLAM receptor on the cells of sheep and goats, initiating cell entry[13]. However, due to small mutations on the SLAM receptor between humans and ruminants, the PPR hemagglutinin cannot bind to human SLAM; preventing PPR from making the jump from animal pathogen to emerging human pathogen.
Emerging pathogens arise when diseases such as PPR make the jump from an animal to a newly exposed human host, with glycoproteins that bind readily to human cell surface receptors, allowing the virus to replicate throughout the human host. These interspecies jumps, to a host with no pre-existing immunity or evolved resistance, result in an unchecked infection before the body can develop an adaptive response against the new pathogen, leading to high mortality rates. I will now consider some of the most dangerous emerging pathogens of the 21st century, avian influenza and Ebola.
Avian Influenza
Avian Flu is one of the most commonly feared emerging pathogens, periodically breaking out in pockets across the globe. Flu has earned this reputation as a consequence of its ubiquity, annually killing up to 650,000 people worldwide[14]. The aforementioned Spanish Flu[1], for example, was one on the most lethal epidemics humanity has ever seen.
Flu is a negative sense strand RNA virus, of the family Orthomyxoviridae, with a genome divided into 8 segments[15]. Specific strains are defined by two surface glycoproteins, hemagglutinin (H) and neuraminidase (N). Alongside native pools of infectious influenza in humans, it is also endemic in bats, birds, pigs, dogs and cats[15]. Within their endemic species, influenza circulates and would not be classed as an emerging pathogen. However, influenza pandemics can occur when the genomes of influenza from different species re-assort[15]. Often using pigs as melting pot[16], re-assortment occurs when two strains of influenza simultaneously infect the same cell (Fig. 3). The two parent viruses proceed to produce capsid proteins and copy their RNA into 8 segments. As the RNA gets packaged, genetic material from different parent viruses can end up in the same capsid, producing a recombinant organism. With co-infection of just two strains there are 256 different possible combinations in which the RNA can be packaged, producing a huge number of variant strains. The result is a virus that can potentially infect a new organism with little pre-existing immunity. Noteworthy examples of this include swine flu[16] (H1N1) and avian flu[17] (H5N1).
Figure 3 Re-assortment of two distinct strains of virus during co-infection. Swine and avian influenza can both infect pig cells. During virion formation strands of RNA from each can be re-assorted into the same capsid, producing a virus with a mixed heritage, better able to infect new targets. Though illustrated with influenza, such re-assortment is extremely common in segmented genome viruses including Rift Valley and Crimean Congo Haemorrhagic Fever; both WHO neglected emerging pathogens.
H5N1, a purely avian influenza with no circulating human strain, appears to have evolved in the late 20th century[17]. It infects many species, including poultry and wild birds[17]. On infection of the former, the close contact with keepers provides sufficient exposure for the virus to make the species jump from birds to humans [17]. Once contracted H5N1 has an estimated 50% mortality rate in humans[18], which would cause a catastrophic pandemic if human-human transmission became commonplace.
Influenza is clearly a dangerous emerging pathogen. The relative mildness of seasonal flu makes populations complacent about the risk. However, combining significant levels of contagiousness with potentially high mortality rates, influenza likely poses the largest risk to global health of any emerging pathogen.
Filoviruses
The filovirus family contains two viral species, Marburgvirus and Ebolavirus; some of the most lethal pathogens known to man. The family contains six species, Marburg virus (MARV) and five species of Ebolavirus. The Zaire, Sudan, Tai Forest and Bundibugyo strains all cause human disease[19], whilst Reston Ebolavirus only infects monkeys [19]. Reston is also unique in that it is native to the Philippines[19]; 10,000Km from Sudan, the closest reservoir of any of the other filoviruses. Here I will focus on the worst filoviruses, the Zaire (up to 90% mortality)[20] and Sudan (up to 65% mortality) strains [20].
The first records of Ebolavirus occur in 1976[21], with the first Sudan outbreak occurring in July (65% mortality rate)[21] and the first Zaire outbreak beginning in late August[22] (88% mortality rate[20]) of that year. Such devastating mortality is one of the hallmarks of emerging pathogens. For Ebolaviruses a challenge has been finding the original animal reservoir. The first transmission of ebolavirus into humans is thought to have come from contact with animals, probably via consumption of bush meat[22]. Though filoviruses appear to have spread to humans from monkeys or apes, we know that they cannot be the natural host, as they also suffer severe mortality from these infections [23][24]. The most likely animal reservoir are bats[25], which are also the reservoir for Marburgvirus; though how they remain infected without symptoms is unknown.
Ebolavirus infection causes severe haemorrhagic fever, as uncontrolled viral replication destroys host cells and leads to systemic infection. The Ebola glycoprotein binds NCP1[27], a cholesterol transporter expressed by many cell types. This gives Ebolavirus the potential to infect a wide range of species and cell types, thus producing simultaneous infections across the body. Particularly targeted are dendritic [28] and endothelial cells[29]. Targeting the former helps Ebola infections evade the adaptive immune response[28], allowing infection to spread unchecked. Specific targeting of endothelial cells causes the symptoms of haemorrhagic fever. As these cells lyse on virus particle exit, blood vessels weaken resulting in bleeding in the GI tract[29], and out of all bodily orifices[29]; a truly horrific series of events leading to rapid death in the majority of infected individuals. Cause of death is hypotension following mass blood loss, leaving the afflicted unable to supply sufficient oxygen to their tissues.
HIV
Human Immunodeficiency Virus (HIV) is one of the most successful emerging pathogens, having spread as a pandemic around the world and now thought to have infected 70 million people[30]; with almost 35 million people dying of AIDS (Acquired Immuno-deficiency Syndrome) in the last 40 years[30]. HIV is part of a family of immunodeficiency viruses, which infect mammals including both cats[31] and non-human primates[32]. The latter appear to be the endemic host of the HIV-related SIV (Simian Immunodeficiency Virus), which circulates in the population, without causing AIDS-like symptoms[32]. At some point in the 1970s, contact between non-primate and human bodily fluids led to transmission into humans[33]; an event only apparent many years later when AIDS was first recognised in 1981[33].
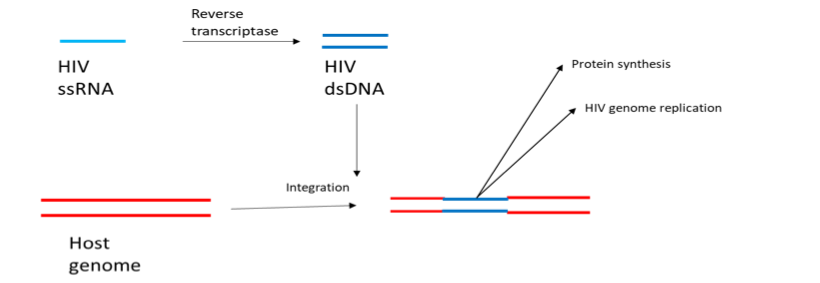
Figure 4 Reverse transcription in the retrovirus life cycle. HIV has a single stranded RNA (ssRNA) genome. Reverse transcriptase, encoded by HIV, copies ssRNA into double stranded DNA (dsDNA) which is then integrated into the host cell genome. From here it can be transcribed back into RNA, facilitating HIV genome replication or protein production.
The HIV-1 M strain is responsible for the continuing pandemic and initiates cell entry through interaction between CD4 and another receptor[34]. The requirement for CD4 binding determines cellular tropism as HIV only infect CD4+ T-cells[34], part of the adaptive immune system. The virus is further split into the two sub-strains depending on the accessory receptor used to initiate cell entry. The R5 strain uses CCR5, whilst X4 uses CXCR4[34]. Post entry, HIV replication is different to that of the previously discussed viruses. HIV is a retrovirus, meaning that in execution of its lifecycle the viral genetic code of the virus is integrated into the host genome[35]. Integration is achieved by reverse transcribing its RNA genome, converting the genome into DNA, which is then inserted into the genome of the host[35] (Fig.4). With HIV DNA within the host cell nucleus the virus remains relatively dormant, producing low levels of virus particles[35] which remain undetected by the immune system.
AIDS arises after many years of such asymptomatic infection, during which HIV can be disseminated without the afflicted knowing they are a carrier. After a period of latency, which can vary widely, the patient progressively develops AIDS[36]; typified by significant depletion in CD4+ T-cells within the immune system. Although CD4+ cells are destroyed in the early stages of infection, pre-AIDS[36], these are regenerated from stem cells, allowing the immune system to continue to function. The ability for the immune system to regenerate degrades with chronic HIV infection[36] however, and eventually the body can no longer replace the lost CD4+ cells, leading to rapid depletion and onset of AIDS[36]. CD4+ cells are a crucial part of the immune system, known as T-helper cells[37]. They manage the rest of the immune system, activating in the case of pathogenic threats[37] and dampening responses against the host’s own proteins[37]. CD4+ cells also modulate the host response depending on the pathogen, activating tailored responses against viruses, bacteria and parasites[37]. Without CD4+ cells to regulate the immune system in AIDS, immune responses cannot be mobilised. The hallmark of AIDS is the presence of secondary opportunistic infections such as the bacterial Pneumocystis carinii, or Mycobacterium tuberculosis[36], viral infections such as herpes simplex or adenovirus[36], or opportunistic cancers such as Kaposi’s sacrcoma[36]; all examples of diseases which would normally be prevented by the mounting of a robust immune response.
Conclusions
To summarise, emerging pathogens are clearly one of the biggest threats to human health. The capacity for explosive outbreaks, akin to Spanish influenza, or for a slow seep into the populace, as seen for HIV, means continued vigilance is essential. Influenza and Ebola both, through unique biological mechanisms, cause severe disease. The latter has emerged into humans repeatedly during the last forty years and appears to show no signs of stopping, causing two outbreaks in the Dominican Republic of Congo this year already. Ebola also devastates ecosystems, highlighting the problem of emerging viruses do not stop at humans. Influenza continuously circulates, giving the population some protection, but re-assortment to display new antigens allows influenza to circumvent natural immunity and cause more severe disease. HIV gives us an example of a pathogen that has emerged relatively recently, and spread rapidly worldwide.
In February 2018 the World Health Organisation published their priority list of pandemic potential emerging diseases[38]. These are:
• Crimean-Congo haemorrhagic fever (CCHF)
• Ebola virus disease and Marburg virus disease
• Lassa fever
• Middle East respiratory syndrome coronavirus (MERS-CoV) and Severe Acute Respiratory Syndrome (SARS)
• Nipah and henipaviral diseases
• Rift Valley fever (RVF)
• Zika
• Disease X (unknown pathogen which may cause severe human epidemics)
These, exclusively viral diseases, are the highest priority for investment in therapeutics and prophylactics to prevent their emergence as epidemics. The diseases on this list share several similarities to those assessed earlier in the article. These diseases, like Ebola and Influenza, circulate in an animal host, producing a continuous lineage and a constant opportunity for mutation to allow re-infection to a non-host organism. On jumping to humans they mostly have high mortality rates, or induce life-altering pathology such as that seen for foetal Zika infections[39]. These diseases, like Ebola, Influenza and HIV before them, thus retain the ability to change the status quo of global public health. On a more optimistic note, steps forward in science are reducing the threat of emerging pathogens. Early warning and identification of these diseases, a luxury not afforded to HIV, alongside decades of vaccine research, honed by the race to keep up with rapidly evolving influenza, is being exploited to generate rapid responses to these new pathogens. Public health programmes, developed in West Africa during the 2014 outbreak, have informed us on how to contain dangerous haemorrhagic fever, spread by blood, for example. Though emerging viral diseases are a major threat to global health, continued scientific advancements prepare us to push back.
References
1. Taubenberger, J. K. (2006). The Origin and Virulence of the 1918 “Spanish” Influenza Virus. Proceedings of the American Philosophical Society, 150(1), 86–112. Retrieved from http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2720273/ [Accessed: 3 May 2018]
2. Mougel, N (2011). World War 1 casualties. REPERES – module 1-0 – explanatory notes – World War I casualties – EN
3. Faust, F. X., Gnecco, C., Mannstein, H., & Stamm, J. (2006). Evidence for the Postconquest Demographic Collapse of the Americas in Historical CO2 Levels. Earth Interactions, 10(11), 1–14. https://doi.org/10.1175/EI157.1
4. Sharp, P. M., & Hahn, B. H. (2011). Origins of HIV and the AIDS Pandemic. Cold Spring Harbor Perspectives in Medicine:, 1(1), a006841. https://doi.org/10.1101/cshperspect.a006841
5. Li, Y. H., & CHEN, S. P. (2014). Evolutionary history of Ebola virus. Epidemiology and Infection, 142(6), 1138–1145. https://doi.org/DOI:10.1017/S0950268813002215
6. Hung, L. S. (2003). The SARS epidemic in Hong Kong: what lessons have we learned? Journal of the Royal Society of Medicine, 96(8), 374–378. Retrieved from http://www.ncbi.nlm.nih.gov/pmc/articles/PMC539564/ [Accessed: 3 May 2018]
7. Tiensin T, Chaitaweesub P, Songserm T, et al. Highly Pathogenic Avian Influenza H5N1, Thailand, 2004. Emerging Infectious Diseases. 2005;11(11):1664-1672. https://doi.org/doi:10.3201/eid1111.050608.
8. Jhung, M. A., Swerdlow, D., Olsen, S. J., Jernigan, D., Biggerstaff, M., Kamimoto, L., … Finelli, L. (2011). Epidemiology of 2009 Pandemic Influenza A (H1N1) in the United States. Clinical Infectious Diseases, 52(suppl_1), S13–S26. Retrieved from http://dx.doi.org/10.1093/cid/ciq008
9. Rossmann, M. G. (2013). Structure of viruses: a short history. Quarterly Reviews of Biophysics, 46(2), 133–180. https://doi.org/DOI:10.1017/S0033583513000012
10. Forterre, P. (2010). Defining Life: The Virus Viewpoint. Origins of Life and Evolution of the Biosphere, 40(2), 151–160. https://doi.org/10.1007/s11084-010-9194-1
11. Banerjee, N., & Mukhopadhyay, S. (2016). Viral glycoproteins: biological role and application in diagnosis. VirusDisease, 27(1), 1–11. https://doi.org/10.1007/s13337-015-0293-5
12. Lin, L.-T., & Richardson, C. D. (2016). The Host Cell Receptors for Measles Virus and Their Interaction with the Viral Hemagglutinin (H) Protein. Viruses, 8(9), 250. https://doi.org/10.3390/v8090250
13. Liang Z, Yuan R, Chen L, Zhu X, Dou Y (2016) Molecular Evolution and Characterization of Hemagglutinin (H) in Peste des Petits Ruminants Virus. PLoS ONE 11(4): e0152587. https://doi.org/10.1371/journal.pone.0152587
14. World Health Organisation URL: http://www.who.int/mediacentre/news/releases/2017/seasonal-flu/en/ [Accessed: 4 May 2018]
15. McDonald, S. M., Nelson, M. I., Turner, P. E., & Patton, J. T. (2016). Reassortment in segmented RNA viruses: mechanisms and outcomes. Nature Reviews. Microbiology, 14(7), 448–460. https://doi.org/10.1038/nrmicro.2016.46.
16. Ducatez, M. F., Hause, B., Stigger-Rosser, E., Darnell, D., Corzo, C., Juleen, K., … Webby, R. J. (2011). Multiple Reassortment between Pandemic (H1N1) 2009 and Endemic Influenza Viruses in Pigs, United States. Emerging Infectious Disease Journal, 17(9), 1624. https://doi.org/10.3201/eid1709.110338
17. Harder, T. C.; Werner, O. (2006). “Avian Influenza”. In Kamps, B. S.; Hoffman, C.; Preiser, W. Influenza Report 2006. Paris, France: Flying Publisher. ISBN 3-924774-51-X. Retrieved 2006-04-18.
18. SEEDMAGAZINE.COM, September 25, 2018, (Originally published January 17, 2006), http://seedmagazine.com/content/article/overestimating_avian_flu/ [Accessed: 4 May 2018]
19. Laupland, K. B., & Valiquette, L. (2014). Ebola virus disease. The Canadian Journal of Infectious Diseases & Medical Microbiology, 25(3), 128–129. Retrieved from http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4173971/ [Accessed: 3 May 2018]
20. World health Organisation Ebolavirus Factsheet URL: http://www.who.int/mediacentre/factsheets/fs103/en/ [Accessed: 3 May 2018]
21. Baron, R. C., McCormick, J. B., & Zubeir, O. A. (1983). Ebola virus disease in southern Sudan: hospital dissemination and intrafamilial spread. Bulletin of the World Health Organization, 61(6), 997–1003. Retrieved from http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2536233/ [Accessed: 4 May 2018]
22. Breman, J. G., Heymann, D. L., Lloyd, G., McCormick, J. B., Miatudila, M., Murphy, F. A., … Johnson, K. M. (2016). Discovery and Description of Ebola Zaire Virus in 1976 and Relevance to the West African Epidemic During 2013–2016. The Journal of Infectious Diseases, 214(suppl_3), S93–S101. Retrieved from http://dx.doi.org/10.1093/infdis/jiw207
23. Miranda, M. E. G., & Miranda, N. L. J. (2011). Reston ebolavirus in Humans and Animals in the Philippines: A Review. The Journal of Infectious Diseases, 204(suppl_3), S757–S760. Retrieved from http://dx.doi.org/10.1093/infdis/jir296
24. Genton C, Cristescu R, Gatti S, Levréro F, Bigot E, Caillaud D, et al. (2012) Recovery Potential of a Western Lowland Gorilla Population following a Major Ebola Outbreak: Results from a Ten Year Study. PLoS ONE 7(5): e37106. https://doi.org/10.1371/journal.pone.0037106
25. Leendertz, S. A. J. (2016). Testing New Hypotheses Regarding Ebolavirus Reservoirs. Viruses, 8(2), 30. https://doi.org/10.3390/v8020030
26. Kuzmin, I. V, Niezgoda, M., Franka, R., Agwanda, B., Markotter, W., Breiman, R. F., … Rupprecht, C. E. (2010). Marburg Virus in Fruit Bat, Kenya. Emerging Infectious Diseases, 16(2), 352–354.https://doi.org/10.3201/eid1602.091269
27. Carette, J. E., Raaben, M., Wong, A. C., Herbert, A. S., Obernosterer, G., Mulherkar, N., … Brummelkamp, T. R. (2011). Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature, 477(7364), 340–343. http://doi.org/10.1038/nature10348
28. Messaoudi, I., Amarasinghe, G. K., & Basler, C. F. (2015). Filovirus pathogenesis and immune evasion: insights from Ebola virus and Marburg virus. Nat Rev Micro, 13(11), 663–676. Retrieved from http://dx.doi.org/10.1038/nrmicro3524
29. Paessler, S., & Walker, D. H. (2013). Pathogenesis of the Viral Hemorrhagic Fevers. Annual Review of Pathology: Mechanisms of Disease, 8(1), 411–440. http://doi.org/10.1146/annurev-pathol-020712-164041
30. WHO HIV factsheet, URL: http://www.who.int/gho/hiv/en/ [Accessed: 3 May 2018]
31. Litster, A. L. (2014). Transmission of feline immunodeficiency virus (FIV) among cohabiting cats in two cat rescue shelters. The Veterinary Journal, 201(2), 184–188. https://doi.org/https://doi.org/10.1016/j.tvjl.2014.02.030
32. Sharp, P. M., Shaw, G. M., & Hahn, B. H. (2005). Simian Immunodeficiency Virus Infection of Chimpanzees. Journal of Virology , 79(7), 3891–3902. https://doi.org/10.1128/JVI.79.7.3891-3902.2005
33. Sharp, P. M., & Hahn, B. H. (2011). Origins of HIV and the AIDS Pandemic. Cold Spring Harbor Perspectives in Medicine:, 1(1), a006841. https://doi.org/10.1101/cshperspect.a006841
34. Wilen, C. B., Tilton, J. C., & Doms, R. W. (2012). HIV: Cell Binding and Entry. Cold Spring Harbor Perspectives in Medicine, 2(8), a006866. https://doi.org/10.1101/cshperspect.a006866
35. Hughes, S. H., & Coffin, J. M. (2016). What Integration Sites Tell Us About HIV Persistence. Cell Host & Microbe, 19(5), 588–598. https://doi.org/10.1016/j.chom.2016.04.010
36. Okoye, A. A., & Picker, L. J. (2013). CD4(+) T cell depletion in HIV infection: mechanisms of immunological failure. Immunological Reviews, 254(1), 54–64. https://doi.org/10.1111/imr.12066
37. Zhu, J., Yamane, H., & Paul, W. E. (2010). Differentiation of Effector CD4 T Cell Populations. Annual Review of Immunology, 28(1), 445–489. https://doi.org/10.1146/annurev-immunol-030409-101212
38. WHO: List of Blueprint priority diseases http://www.who.int/blueprint/priority-diseases/en/ [Accessed: 4 May 2018]
39. de Araújo, T. V. B., Ximenes, R. A. de A., Miranda-Filho, D. de B., Souza, W. V., Montarroyos, U. R., de Melo, A. P. L., … Felix Oliveira, V. (2018). Association between microcephaly, Zika virus infection, and other risk factors in Brazil: final report of a case-control study. The Lancet Infectious Diseases, 18(3), 328–336. https://doi.org/10.1016/S1473-3099(17)30727-2

Emerging Viral Pathogens: New Threats in Global Health by Ciaran Gilbride is licensed under a Creative Commons Attribution 4.0 International License.
<< Back to Contents
<< Back to Publications
St Anne's Academic Review (STAAR) A Publication by St Anne's College Middle Common Room ISSN 2048-2566 (Online) ISSN 2515-6527 (Print)


Leave a Reply
You must be logged in to post a comment.